Analysis Summary
This report aggregates results from rMAP modules (QC, assembly/annotation, pangenome/phylogeny, AMR/MGE/virulence/BLAST, and phylogeny).
Quality Control
Per-sample QC reports
- QC Report 1 — SRR8948882
- QC Report 2 — SRR8948883
- QC Report 3 — SRR8948883
- QC Report 4 — SRR8948884
- QC Report 5 — SRR8948884
- QC Report 6 — SRR8948885
- QC Report 7 — SRR8948885
- QC Report 8 — SRR8948886
- QC Report 9 — SRR8948886
- QC Report 10 — SRR8948887
- QC Report 11 — SRR8948878
- QC Report 12 — SRR8948887
- QC Report 13 — SRR8948888
- QC Report 14 — SRR8948888
- QC Report 15 — SRR8948889
- QC Report 16 — SRR8948889
- QC Report 17 — SRR8948890
- QC Report 18 — SRR8948890
- QC Report 19 — SRR8948891
- QC Report 20 — SRR8948891
- QC Report 21 — SRR8948878
- QC Report 22 — SRR8948879
- QC Report 23 — SRR8948879
- QC Report 24 — SRR8948880
- QC Report 25 — SRR8948880
- QC Report 26 — SRR8948881
- QC Report 27 — SRR8948881
- QC Report 28 — SRR8948882
Read trimming
| Sample | Input Pairs | Both Surviving | Forward Only | Reverse Only | Dropped (%) | Survival Rate (%) |
|---|---|---|---|---|---|---|
| SRR8948878 | 798695 | 686272 | 89774 | 6881 | 1.97% | 98.03% |
| SRR8948879 | 795565 | 704552 | 68196 | 8554 | 1.79% | 98.21% |
| SRR8948880 | 798476 | 707768 | 68715 | 8166 | 1.73% | 98.27% |
| SRR8948881 | 796777 | 702646 | 73041 | 7542 | 1.70% | 98.30% |
| SRR8948882 | 794677 | 690974 | 82136 | 7037 | 1.83% | 98.17% |
| SRR8948883 | 791155 | 686762 | 81358 | 7678 | 1.94% | 98.06% |
| SRR8948884 | 797008 | 708498 | 67058 | 8099 | 1.68% | 98.32% |
| SRR8948885 | 793703 | 698515 | 72734 | 8129 | 1.80% | 98.20% |
| SRR8948886 | 793366 | 698501 | 71982 | 8190 | 1.85% | 98.15% |
| SRR8948887 | 794174 | 698245 | 73190 | 8193 | 1.83% | 98.17% |
| SRR8948888 | 789960 | 699891 | 67503 | 8501 | 1.78% | 98.22% |
| SRR8948889 | 796379 | 709445 | 65106 | 8466 | 1.68% | 98.32% |
| SRR8948890 | 795517 | 688686 | 83627 | 7485 | 1.98% | 98.02% |
| SRR8948891 | 796160 | 702080 | 71553 | 8133 | 1.81% | 98.19% |
Assembly
Assembly statistics
| Sample | Contigs | Total Length | Min Length | Max Length | Avg Length | N50 |
|---|---|---|---|---|---|---|
| SRR8948878 | 12613 | 5386819 | 78 | 273502 | 427 | 16861 |
| SRR8948879 | 8758 | 5238305 | 78 | 245309 | 598 | 18589 |
| SRR8948880 | 8620 | 5491052 | 78 | 273483 | 637 | 15660 |
| SRR8948881 | 2674 | 3287057 | 78 | 200750 | 1229 | 63261 |
| SRR8948882 | 2778 | 3293639 | 78 | 113420 | 1185 | 22403 |
| SRR8948883 | 1803 | 3197525 | 78 | 93483 | 1773 | 23568 |
| SRR8948884 | 14662 | 7806957 | 78 | 200750 | 532 | 908 |
| SRR8948885 | 1828 | 3615890 | 78 | 423875 | 1978 | 109187 |
| SRR8948886 | 6982 | 3905010 | 78 | 173043 | 559 | 34899 |
| SRR8948887 | 3750 | 5856934 | 78 | 245309 | 1561 | 7931 |
| SRR8948888 | 2403 | 3330695 | 78 | 369354 | 1386 | 62414 |
| SRR8948889 | 2972 | 6255193 | 78 | 272949 | 2104 | 37601 |
| SRR8948890 | 3344 | 3475065 | 78 | 369354 | 1039 | 60951 |
| SRR8948891 | 9867 | 6334874 | 78 | 247312 | 642 | 1614 |
Annotation
Annotation summary
| Sample ID | Gene Count |
|---|---|
| SRR8948878 | 3376 |
| SRR8948879 | 3675 |
| SRR8948880 | 3992 |
| SRR8948881 | 2745 |
| SRR8948882 | 2812 |
| SRR8948883 | 2871 |
| SRR8948884 | 5270 |
| SRR8948885 | 3257 |
| SRR8948886 | 2747 |
| SRR8948887 | 4996 |
| SRR8948888 | 2893 |
| SRR8948889 | 5635 |
| SRR8948890 | 2899 |
| SRR8948891 | 4627 |
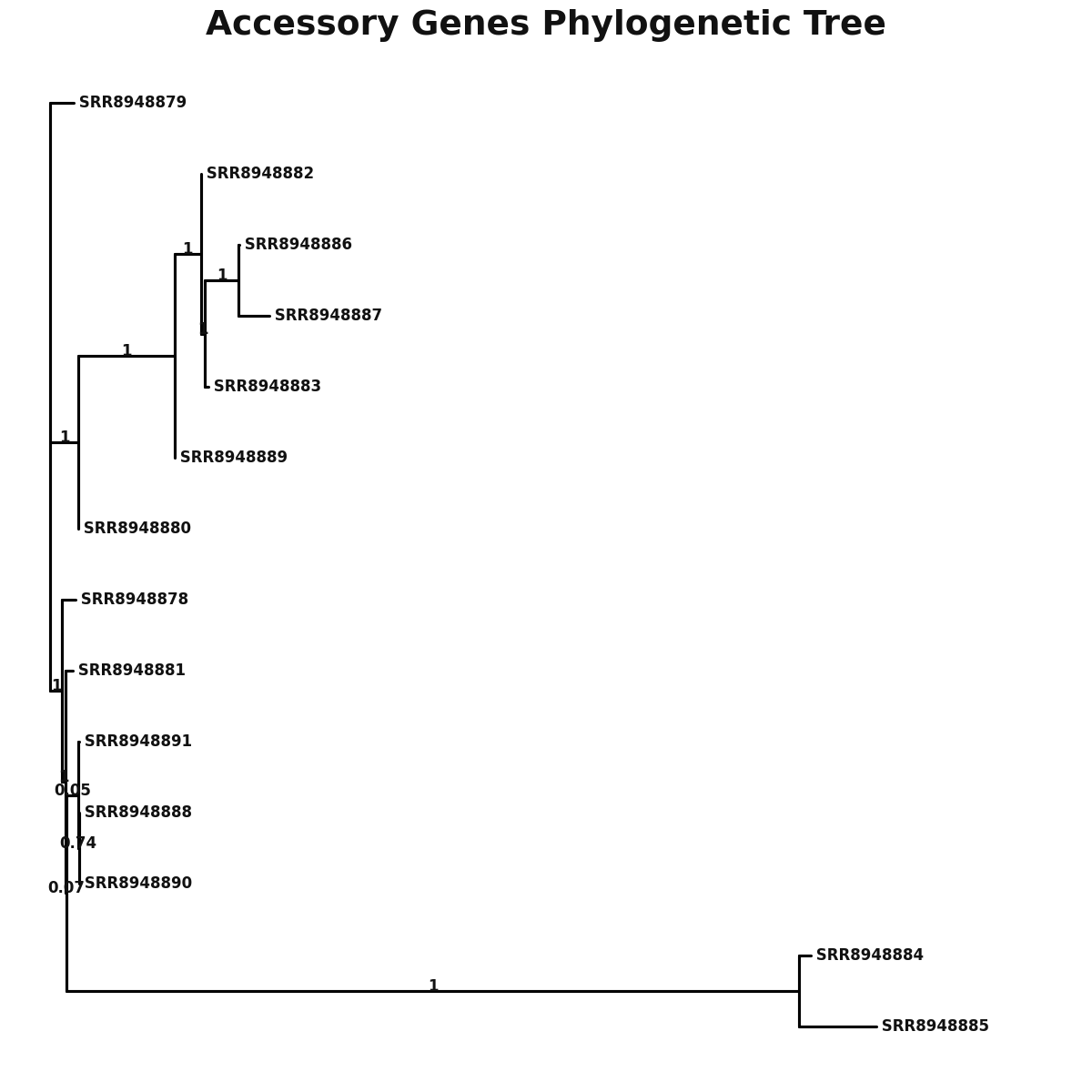
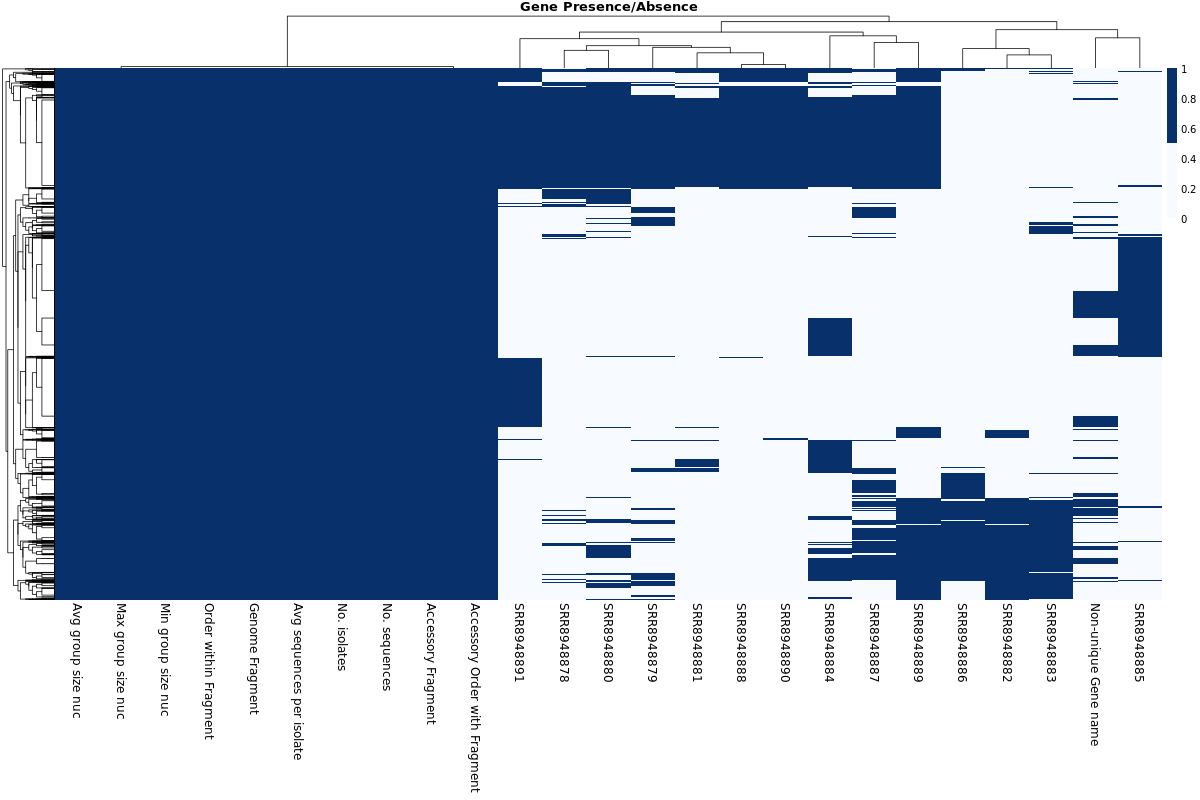
Pangenome
Pangenome report
Pangenome Analysis
Roary completed successfully. (This page will show Roary results if available.)

Gene Presence/Absence Heatmap
MLST
Combined MLST
| Sample ID | Species | ST | gapA | infB | mdh | pgi | phoE | rpoB | tonB |
|---|---|---|---|---|---|---|---|---|---|
| SRR8948878 | efaecalis | 16 | 5 | 1 | 1 | 3 | 7 | 7 | 6 |
| SRR8948879 | efaecalis | 40 | 3 | 6 | 23 | 12 | 9 | 10 | 7 |
| SRR8948880 | efaecalis | 16 | 5 | 1 | 1 | 3 | 7 | 7 | 6 |
| SRR8948881 | efaecalis | 40 | 3 | 6 | 23 | 12 | 9 | 10 | 7 |
| SRR8948882 | efaecium | 80 | 9 | 1 | 1 | 1 | 12 | 1 | 1 |
| SRR8948883 | efaecium | 203 | 15 | 1 | 1 | 1 | 1 | 20 | 1 |
| SRR8948884 | efaecalis | - | 3 | 6,11 | 23 | 12 | 9 | 10 | 7 |
| SRR8948885 | - | - | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| SRR8948886 | efaecium | 1446 | 11 | 13 | 18 | 15 | 10 | 19 | 6 |
| SRR8948887 | efaecalis | 40 | 3 | 6 | 23 | 12 | 9 | 10 | 7 |
| SRR8948888 | efaecalis | 179 | 5 | 1 | 1 | 3 | 7 | 1 | 6 |
| SRR8948889 | efaecalis | - | 5,92 | 1 | 1 | 3 | 7 | 1 | 6 |
| SRR8948890 | efaecalis | 179 | 5 | 1 | 1 | 3 | 7 | 1 | 6 |
| SRR8948891 | efaecalis | 179 | 5 | 1 | 1 | 3 | 7 | 1 | 6 |
Variants
Variant summary
| Sample | Status | SNPs | INDELs | Total Variants | Attempts |
|---|---|---|---|---|---|
| SRR8948878 | Success | 16001 | 0 | 16001 | 1 |
| SRR8948879 | Success | 15709 | 0 | 15709 | 1 |
| SRR8948880 | Success | 16025 | 0 | 16025 | 1 |
| SRR8948881 | Success | 15976 | 0 | 15976 | 1 |
| SRR8948882 | Success | 1587 | 0 | 1587 | 1 |
| SRR8948883 | Success | 1617 | 0 | 1617 | 1 |
| SRR8948884 | Success | 15981 | 0 | 15981 | 1 |
| SRR8948885 | Success | 1488 | 0 | 1488 | 1 |
| SRR8948886 | Success | 1451 | 0 | 1451 | 1 |
| SRR8948887 | Success | 15718 | 0 | 15718 | 1 |
| SRR8948888 | Success | 16872 | 0 | 16872 | 1 |
| SRR8948889 | Success | 16709 | 0 | 16709 | 1 |
| SRR8948890 | Success | 16877 | 0 | 16877 | 1 |
| SRR8948891 | Success | 16868 | 0 | 16868 | 1 |
Antimicrobial Resistance
Per-sample AMR reports
- AMR Report 1 — SRR8948878
- AMR Report 2 — SRR8948879
- AMR Report 3 — SRR8948880
- AMR Report 4 — SRR8948881
- AMR Report 5 — SRR8948882
- AMR Report 6 — SRR8948883
- AMR Report 7 — SRR8948884
- AMR Report 8 — SRR8948885
- AMR Report 9 — SRR8948886
- AMR Report 10 — SRR8948887
- AMR Report 11 — SRR8948888
- AMR Report 12 — SRR8948889
- AMR Report 13 — SRR8948890
- AMR Report 14 — SRR8948891
Mobile Genetic Elements
Per-sample MGE reports
- MGE Report 1 — SRR8948878
- MGE Report 2 — SRR8948879
- MGE Report 3 — SRR8948880
- MGE Report 4 — SRR8948881
- MGE Report 5 — SRR8948882
- MGE Report 6 — SRR8948883
- MGE Report 7 — SRR8948884
- MGE Report 8 — SRR8948885
- MGE Report 9 — SRR8948886
- MGE Report 10 — SRR8948887
- MGE Report 11 — SRR8948888
- MGE Report 12 — SRR8948889
- MGE Report 13 — SRR8948890
- MGE Report 14 — SRR8948891
Virulence
Per-sample Virulence reports
- Virulence Report 1 — SRR8948887
- Virulence Report 2 — SRR8948888
- Virulence Report 3 — SRR8948889
- Virulence Report 4 — SRR8948890
- Virulence Report 5 — SRR8948891
- Virulence Report 6 — SRR8948878
- Virulence Report 7 — SRR8948879
- Virulence Report 8 — SRR8948880
- Virulence Report 9 — SRR8948881
- Virulence Report 10 — SRR8948882
- Virulence Report 11 — SRR8948883
- Virulence Report 12 — SRR8948884
- Virulence Report 13 — SRR8948885
- Virulence Report 14 — SRR8948886
BLAST
Per-sample BLAST reports
- BLAST Report 1 — SRR8948878
- BLAST Report 2 — SRR8948879
- BLAST Report 3 — SRR8948880
- BLAST Report 4 — SRR8948881
- BLAST Report 5 — SRR8948882
- BLAST Report 6 — SRR8948883
- BLAST Report 7 — SRR8948884
- BLAST Report 8 — SRR8948885
- BLAST Report 9 — SRR8948886
- BLAST Report 10 — SRR8948887
- BLAST Report 11 — SRR8948888
- BLAST Report 12 — SRR8948889
- BLAST Report 13 — SRR8948890
- BLAST Report 14 — SRR8948891
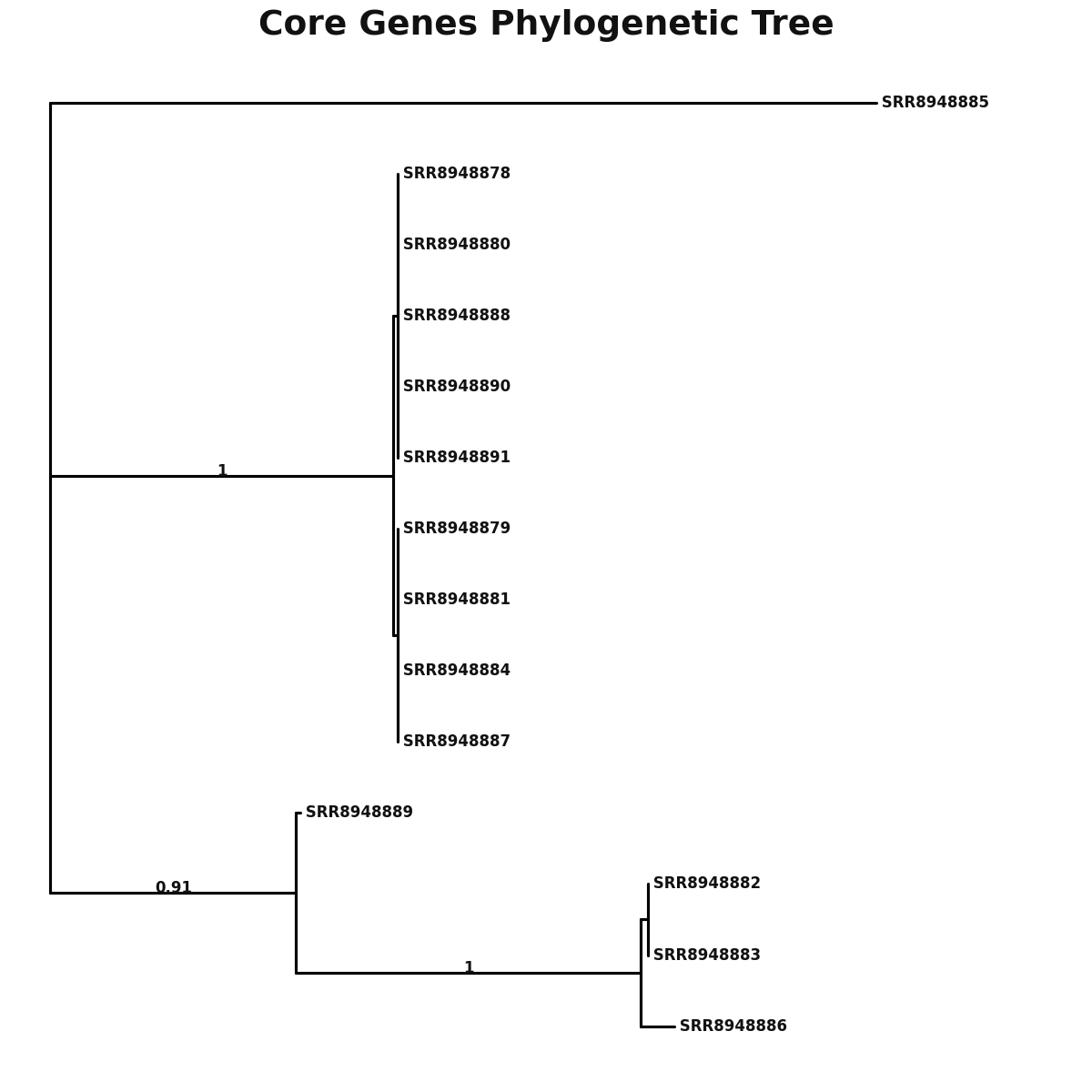
Phylogeny
core_core_genes.png
accessory_accessory_genes.png