Analysis Summary
This report aggregates results from rMAP modules (QC, assembly/annotation, pangenome/phylogeny, AMR/MGE/virulence/BLAST, and phylogeny).
Quality Control
Per-sample QC reports
- QC Report 1 — SRR15793220
- QC Report 2 — SRR16873817
- QC Report 3 — SRR16873817
- QC Report 4 — SRR22839873
- QC Report 5 — SRR22839873
- QC Report 6 — SRR22839890
- QC Report 7 — SRR22839890
- QC Report 8 — SRR22839907
- QC Report 9 — SRR22839907
- QC Report 10 — SRR22839978
- QC Report 11 — ERR1663005
- QC Report 12 — SRR22839978
- QC Report 13 — SRR8640965
- QC Report 14 — SRR8640965
- QC Report 15 — ERR1663005
- QC Report 16 — ERR1664565
- QC Report 17 — ERR1664565
- QC Report 18 — SRR15793157
- QC Report 19 — SRR15793157
- QC Report 20 — SRR15793168
- QC Report 21 — SRR15793168
- QC Report 22 — SRR15793220
Read trimming
| Sample | Input Pairs | Both Surviving | Forward Only | Reverse Only | Dropped (%) | Survival Rate (%) |
|---|---|---|---|---|---|---|
| SRR22839978 | 1733236 | 1539372 | 92518 | 48277 | 3.06% | 96.94% |
| SRR16873817 | 3883701 | 3556024 | 90815 | 43054 | 4.99% | 95.01% |
| ERR1663005 | 539052 | 494460 | 33442 | 4857 | 1.17% | 98.83% |
| SRR8640965 | 973419 | 869986 | 80535 | 9476 | 1.38% | 98.62% |
| SRR15793157 | 2464419 | 2255828 | 74789 | 104867 | 1.17% | 98.83% |
| SRR15793168 | 2241086 | 2039018 | 81579 | 91276 | 1.30% | 98.70% |
| SRR22839907 | 1514440 | 1345412 | 78938 | 42962 | 3.11% | 96.89% |
| ERR1664565 | 204975 | 191700 | 9230 | 1502 | 1.24% | 98.76% |
| SRR15793220 | 2386093 | 2312984 | 30477 | 37012 | 0.24% | 99.76% |
| SRR22839873 | 1626522 | 1439093 | 71335 | 72830 | 2.66% | 97.34% |
| SRR22839890 | 1098592 | 970463 | 59019 | 32532 | 3.33% | 96.67% |
Assembly
Assembly statistics
| Sample | Contigs | Total Length | Min Length | Max Length | Avg Length | N50 |
|---|---|---|---|---|---|---|
| ERR1663005 | 1151 | 7066360 | 78 | 193037 | 6139 | 66477 |
| ERR1664565 | 2974 | 7074209 | 78 | 72893 | 2378 | 8169 |
| SRR15793157 | 4041 | 7315939 | 78 | 282665 | 1810 | 138268 |
| SRR15793168 | 2712 | 6670690 | 78 | 564863 | 2459 | 127847 |
| SRR15793220 | 10830 | 8229645 | 78 | 407884 | 759 | 106356 |
| SRR16873817 | 17265 | 8679160 | 78 | 196798 | 502 | 25098 |
| SRR22839873 | 2318 | 6798706 | 78 | 303916 | 2933 | 102243 |
| SRR22839890 | 1370 | 6389408 | 78 | 290392 | 4663 | 87675 |
| SRR22839907 | 1867 | 6584964 | 78 | 415271 | 3527 | 109075 |
| SRR22839978 | 1714 | 6965962 | 78 | 370288 | 4064 | 91698 |
| SRR8640965 | 875 | 6562170 | 78 | 192831 | 7499 | 84056 |
Annotation
Annotation summary
| Sample ID | Gene Count |
|---|---|
| ERR1663005 | 6457 |
| ERR1664565 | 6436 |
| SRR15793157 | 6331 |
| SRR15793168 | 5818 |
| SRR15793220 | 5952 |
| SRR16873817 | 6289 |
| SRR22839873 | 5952 |
| SRR22839890 | 5632 |
| SRR22839907 | 5806 |
| SRR22839978 | 6211 |
| SRR8640965 | 5887 |
Pangenome
Pangenome report
Pangenome Analysis
Roary completed successfully. (This page will show Roary results if available.)

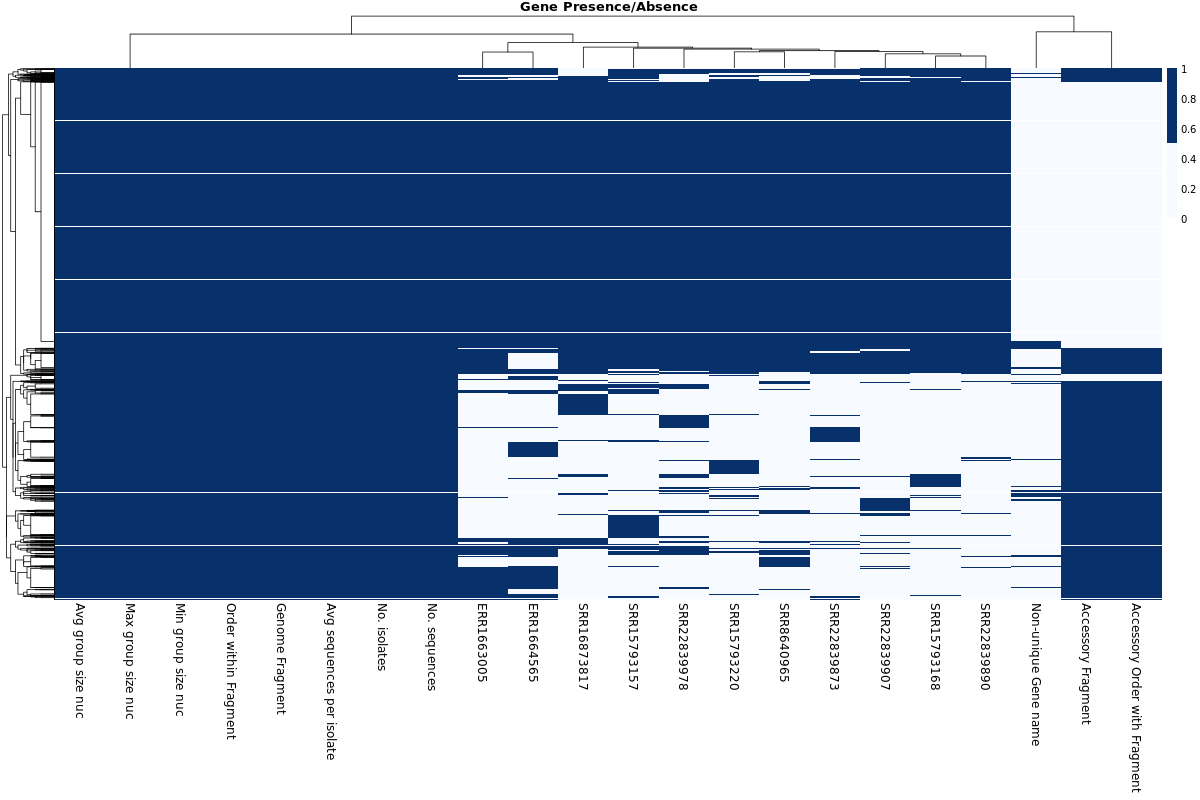
Gene Presence/Absence Heatmap
MLST
Combined MLST
| Sample ID | Species | ST | gapA | infB | mdh | pgi | phoE | rpoB | tonB |
|---|---|---|---|---|---|---|---|---|---|
| ERR1663005 | paeruginosa | 175 | 28 | 22 | 5 | 3 | 3 | 14 | 19 |
| ERR1664565 | paeruginosa | 175 | 28 | 22 | 5 | 3 | 3 | 14 | 19 |
| SRR15793157 | paeruginosa | 17 | 11 | 5 | 1 | 7 | 9 | 4 | 7 |
| SRR15793168 | paeruginosa | 274 | 23 | 5 | 11 | 7 | 1 | 12 | 7 |
| SRR15793220 | paeruginosa | 267 | 19 | 5 | 12 | 11 | 11 | 4 | 14 |
| SRR16873817 | paeruginosa | 395 | 6 | 5 | 1 | 1 | 1 | 12 | 1 |
| SRR22839873 | paeruginosa | 1435 | 16 | 4 | 26 | 3 | 4 | 120 | 1 |
| SRR22839890 | paeruginosa | 1596 | 39 | 3 | 1 | 3 | 3 | 4 | 1 |
| SRR22839907 | paeruginosa | 2100 | 17 | 8 | 3 | 11 | 1 | 15 | 1 |
| SRR22839978 | paeruginosa | 298 | 18 | 4 | 13 | 3 | 1 | 17 | 13 |
| SRR8640965 | paeruginosa | 253 | 4 | 4 | 16 | 12 | 1 | 6 | 3 |
Variants
Variant summary
| Sample | Status | SNPs | INDELs | Total Variants | Attempts |
|---|---|---|---|---|---|
| ERR1663005 | Success | 23018 | 0 | 23018 | 1 |
| ERR1664565 | Success | 10859 | 0 | 10859 | 1 |
| SRR15793157 | Success | 23528 | 0 | 23528 | 1 |
| SRR15793168 | Success | 23546 | 0 | 23546 | 1 |
| SRR15793220 | Success | 18901 | 0 | 18901 | 1 |
| SRR16873817 | Success | 23530 | 0 | 23530 | 1 |
| SRR22839873 | Success | 27235 | 0 | 27235 | 1 |
| SRR22839890 | Success | 22563 | 0 | 22563 | 1 |
| SRR22839907 | Success | 27093 | 0 | 27093 | 1 |
| SRR22839978 | Success | 45902 | 0 | 45902 | 1 |
| SRR8640965 | Success | 45718 | 0 | 45718 | 1 |
Antimicrobial Resistance
Per-sample AMR reports
Mobile Genetic Elements
Per-sample MGE reports
Virulence
Per-sample Virulence reports
- Virulence Report 1 — SRR22839978
- Virulence Report 2 — SRR8640965
- Virulence Report 3 — ERR1663005
- Virulence Report 4 — ERR1664565
- Virulence Report 5 — SRR15793157
- Virulence Report 6 — SRR15793168
- Virulence Report 7 — SRR15793220
- Virulence Report 8 — SRR16873817
- Virulence Report 9 — SRR22839873
- Virulence Report 10 — SRR22839890
- Virulence Report 11 — SRR22839907
BLAST
Per-sample BLAST reports
- BLAST Report 1 — ERR1663005
- BLAST Report 2 — ERR1664565
- BLAST Report 3 — SRR15793157
- BLAST Report 4 — SRR15793168
- BLAST Report 5 — SRR15793220
- BLAST Report 6 — SRR16873817
- BLAST Report 7 — SRR22839873
- BLAST Report 8 — SRR22839890
- BLAST Report 9 — SRR22839907
- BLAST Report 10 — SRR22839978
- BLAST Report 11 — SRR8640965
Phylogeny
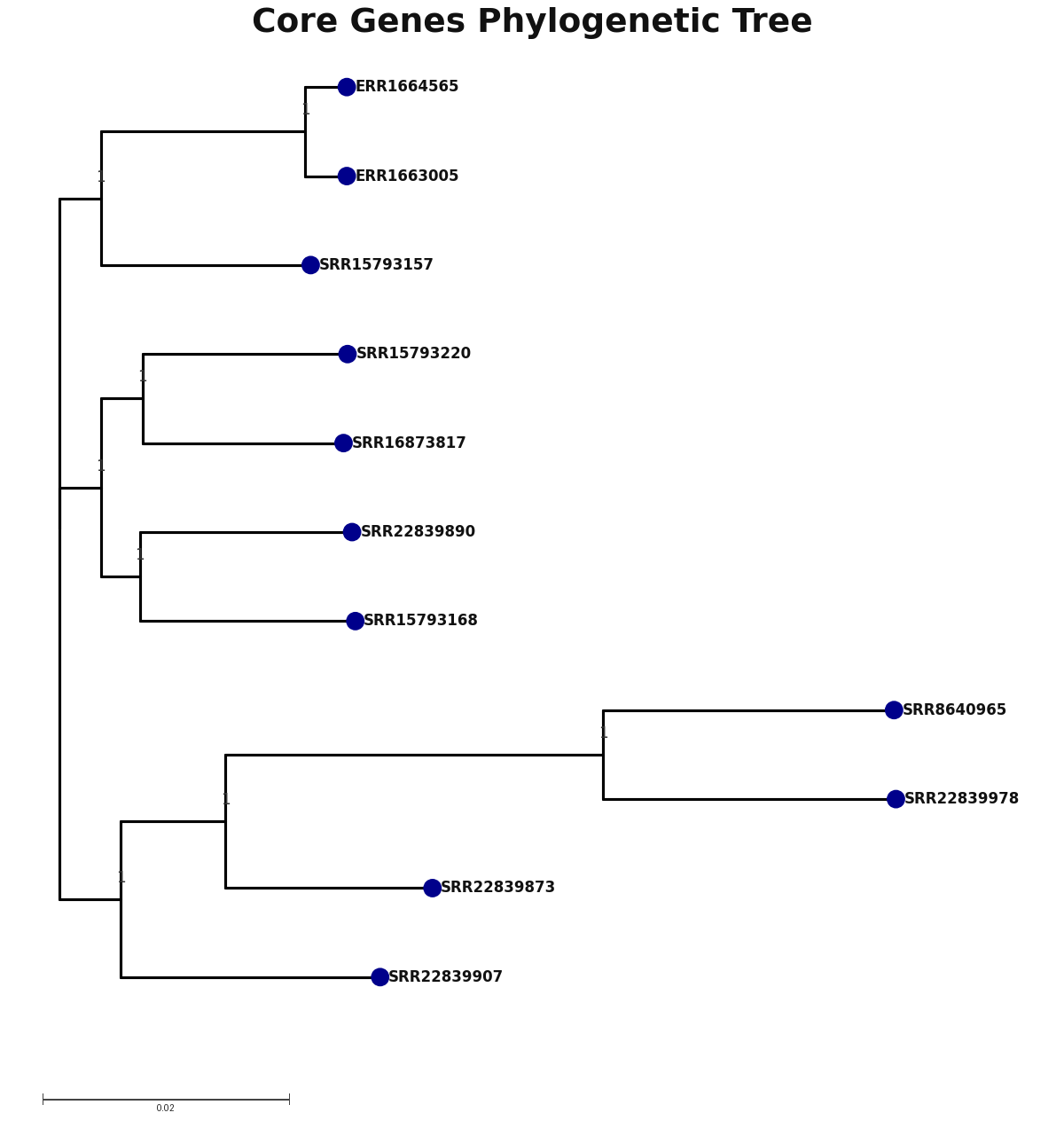
core_core_genes.png
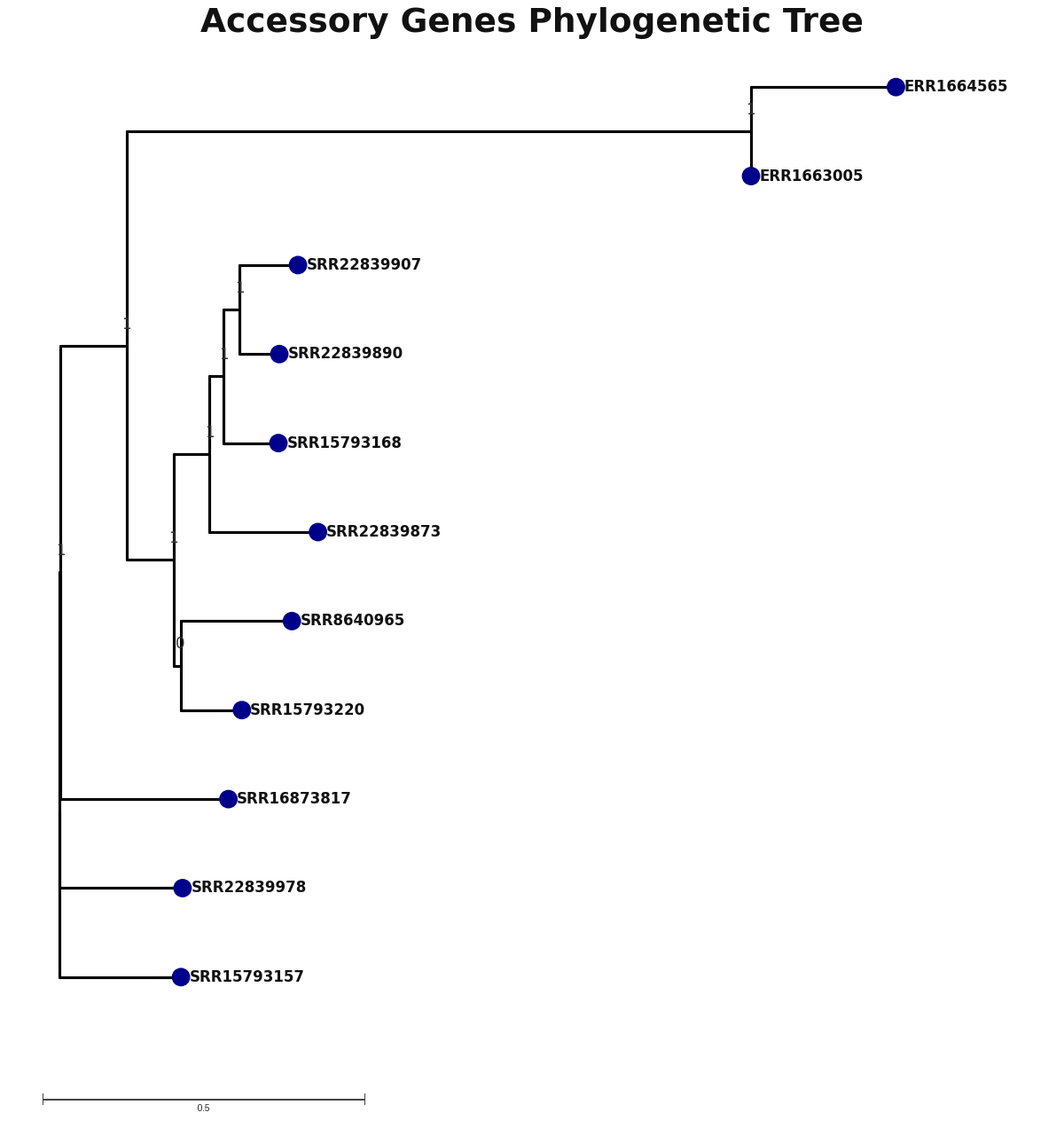
accessory_accessory_genes.png