Analysis Summary
This report aggregates results from rMAP modules (QC, assembly/annotation, pangenome/phylogeny, AMR/MGE/virulence/BLAST, and phylogeny).
Quality Control
Per-sample QC reports
- QC Report 1 — ERR1794904
- QC Report 2 — ERR1794905
- QC Report 3 — ERR1794905
- QC Report 4 — ERR1794906
- QC Report 5 — ERR1794906
- QC Report 6 — ERR1794907
- QC Report 7 — ERR1794907
- QC Report 8 — ERR1794908
- QC Report 9 — ERR1794908
- QC Report 10 — ERR1794909
- QC Report 11 — ERR1794900
- QC Report 12 — ERR1794909
- QC Report 13 — ERR1794910
- QC Report 14 — ERR1794910
- QC Report 15 — ERR1794911
- QC Report 16 — ERR1794911
- QC Report 17 — ERR1794912
- QC Report 18 — ERR1794912
- QC Report 19 — ERR1794913
- QC Report 20 — ERR1794913
- QC Report 21 — ERR1794914
- QC Report 22 — ERR1794900
- QC Report 23 — ERR1794914
- QC Report 24 — ERR1794901
- QC Report 25 — ERR1794901
- QC Report 26 — ERR1794902
- QC Report 27 — ERR1794902
- QC Report 28 — ERR1794903
- QC Report 29 — ERR1794903
- QC Report 30 — ERR1794904
Read trimming
| Sample | Input Pairs | Both Surviving | Forward Only | Reverse Only | Dropped (%) | Survival Rate (%) |
|---|---|---|---|---|---|---|
| ERR1794904 | 512997 | 494352 | 13274 | 2770 | 0.51% | 99.49% |
| ERR1794909 | 2803887 | 2537579 | 96452 | 68201 | 3.63% | 96.37% |
| ERR1794910 | 1211509 | 1188214 | 4462 | 10668 | 0.67% | 99.33% |
| ERR1794901 | 1725334 | 1600498 | 100888 | 5253 | 1.08% | 98.92% |
| ERR1794906 | 718383 | 691238 | 19973 | 3240 | 0.55% | 99.45% |
| ERR1794912 | 1237270 | 1157427 | 63909 | 4940 | 0.89% | 99.11% |
| ERR1794903 | 1557916 | 1476179 | 54858 | 12009 | 0.95% | 99.05% |
| ERR1794911 | 747947 | 723959 | 15264 | 4489 | 0.57% | 99.43% |
| ERR1794900 | 420907 | 388061 | 27942 | 1448 | 0.82% | 99.18% |
| ERR1794905 | 593319 | 573395 | 13301 | 3291 | 0.56% | 99.44% |
| ERR1794908 | 864905 | 829639 | 26019 | 3920 | 0.62% | 99.38% |
| ERR1794914 | 2277997 | 2109538 | 48844 | 67143 | 2.30% | 97.70% |
| ERR1794913 | 626224 | 605565 | 12893 | 3883 | 0.62% | 99.38% |
| ERR1794902 | 1860449 | 1777864 | 53252 | 13682 | 0.84% | 99.16% |
| ERR1794907 | 797659 | 771917 | 15669 | 5263 | 0.60% | 99.40% |
Assembly
Assembly statistics
| Sample | Contigs | Total Length | Min Length | Max Length | Avg Length | N50 |
|---|---|---|---|---|---|---|
| ERR1794900 | 389 | 2782493 | 78 | 145107 | 7152 | 42291 |
| ERR1794901 | 322 | 2740640 | 78 | 180681 | 8511 | 69912 |
| ERR1794902 | 376 | 2793056 | 78 | 162843 | 7428 | 64471 |
| ERR1794903 | 365 | 2786462 | 78 | 141683 | 7634 | 64093 |
| ERR1794904 | 364 | 2740590 | 78 | 104449 | 7529 | 53879 |
| ERR1794905 | 306 | 2741029 | 78 | 147361 | 8957 | 57559 |
| ERR1794906 | 310 | 2739412 | 78 | 140672 | 8836 | 62605 |
| ERR1794907 | 582 | 2771172 | 78 | 173930 | 4761 | 57563 |
| ERR1794908 | 528 | 2777875 | 78 | 159792 | 5261 | 47143 |
| ERR1794909 | 487 | 2775693 | 78 | 206416 | 5699 | 45928 |
| ERR1794910 | 512 | 2772832 | 78 | 159842 | 5415 | 45178 |
| ERR1794911 | 503 | 2879448 | 78 | 193969 | 5724 | 51188 |
| ERR1794912 | 802 | 2859866 | 78 | 252044 | 3565 | 57418 |
| ERR1794913 | 464 | 2831981 | 78 | 175348 | 6103 | 59502 |
| ERR1794914 | 635 | 2846121 | 78 | 150950 | 4482 | 62611 |
Annotation
Annotation summary
| Sample ID | Gene Count |
|---|---|
| ERR1794900 | 2554 |
| ERR1794901 | 2499 |
| ERR1794902 | 2565 |
| ERR1794903 | 2553 |
| ERR1794904 | 2494 |
| ERR1794905 | 2498 |
| ERR1794906 | 2499 |
| ERR1794907 | 2498 |
| ERR1794908 | 2507 |
| ERR1794909 | 2515 |
| ERR1794910 | 2507 |
| ERR1794911 | 2620 |
| ERR1794912 | 2583 |
| ERR1794913 | 2596 |
| ERR1794914 | 2567 |
Pangenome
Pangenome report
Pangenome Analysis
Roary completed successfully. (This page will show Roary results if available.)

Gene Presence/Absence Heatmap
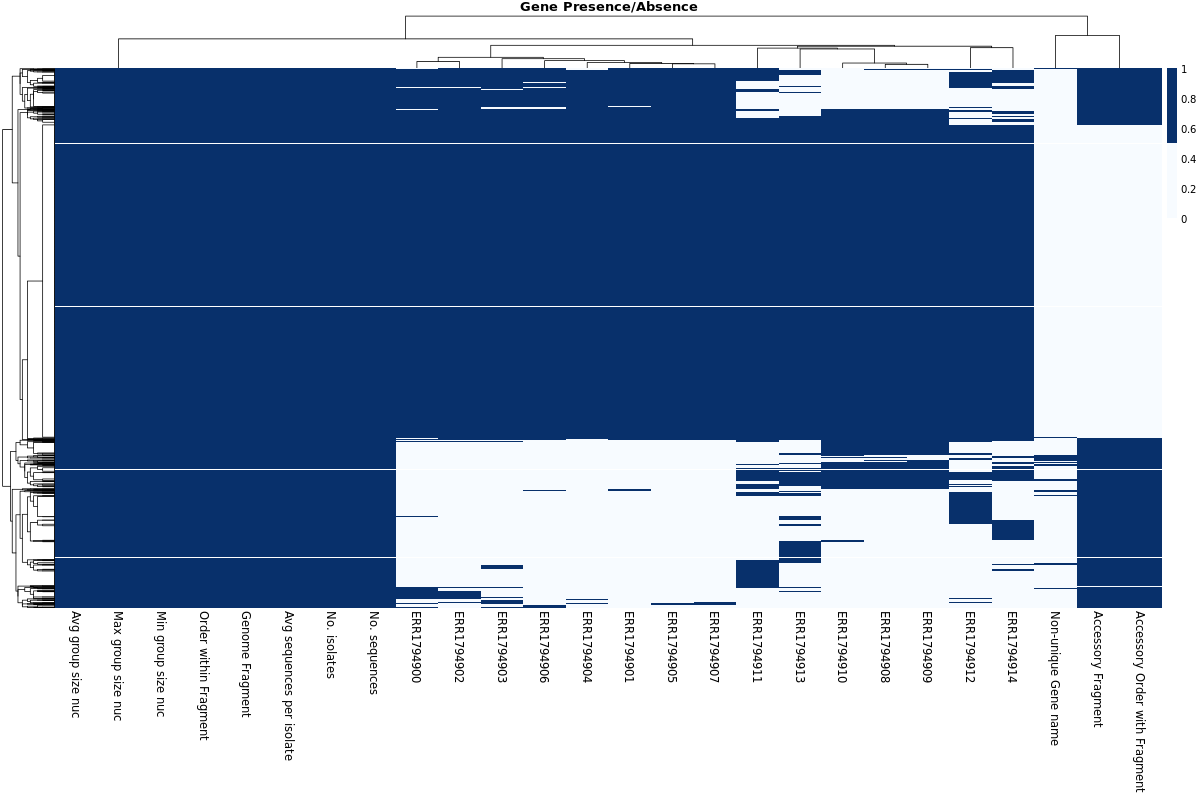
MLST
Combined MLST
| Sample ID | Species | ST | gapA | infB | mdh | pgi | phoE | rpoB | tonB |
|---|---|---|---|---|---|---|---|---|---|
| ERR1794900 | saureus | 152 | 46 | 75 | 49 | 44 | 13 | 68 | 60 |
| ERR1794901 | saureus | 152 | 46 | 75 | 49 | 44 | 13 | 68 | 60 |
| ERR1794902 | saureus | 152 | 46 | 75 | 49 | 44 | 13 | 68 | 60 |
| ERR1794903 | saureus | 152 | 46 | 75 | 49 | 44 | 13 | 68 | 60 |
| ERR1794904 | saureus | 152 | 46 | 75 | 49 | 44 | 13 | 68 | 60 |
| ERR1794905 | saureus | 152 | 46 | 75 | 49 | 44 | 13 | 68 | 60 |
| ERR1794906 | saureus | 152 | 46 | 75 | 49 | 44 | 13 | 68 | 60 |
| ERR1794907 | saureus | 1633 | 46 | 75 | 212 | 44 | 13 | 68 | 60 |
| ERR1794908 | saureus | 15 | 13 | 13 | 1 | 1 | 12 | 11 | 13 |
| ERR1794909 | saureus | 15 | 13 | 13 | 1 | 1 | 12 | 11 | 13 |
| ERR1794910 | saureus | 15 | 13 | 13 | 1 | 1 | 12 | 11 | 13 |
| ERR1794911 | saureus | 72 | 1 | 4 | 1 | 8 | 4 | 4 | 3 |
| ERR1794912 | saureus | 121 | 6 | 5 | 6 | 2 | 7 | 14 | 5 |
| ERR1794913 | saureus | 5 | 1 | 4 | 1 | 4 | 12 | 1 | 10 |
| ERR1794914 | saureus | 80 | 1 | 3 | 1 | 14 | 11 | 51 | 10 |
Variants
Variant summary
| Sample | Status | SNPs | INDELs | Total Variants | Attempts |
|---|---|---|---|---|---|
| ERR1794900 | Success | 33795 | 0 | 33795 | 1 |
| ERR1794901 | Success | 37168 | 0 | 37168 | 1 |
| ERR1794902 | Success | 37091 | 0 | 37091 | 1 |
| ERR1794903 | Success | 37081 | 0 | 37081 | 1 |
| ERR1794904 | Success | 34895 | 0 | 34895 | 1 |
| ERR1794905 | Success | 35575 | 0 | 35575 | 1 |
| ERR1794906 | Success | 36087 | 0 | 36087 | 1 |
| ERR1794907 | Success | 36175 | 0 | 36175 | 1 |
| ERR1794908 | Success | 31062 | 0 | 31062 | 1 |
| ERR1794909 | Success | 31502 | 0 | 31502 | 1 |
| ERR1794910 | Success | 31300 | 0 | 31300 | 1 |
| ERR1794911 | Success | 30851 | 0 | 30851 | 1 |
| ERR1794912 | Success | 28211 | 0 | 28211 | 1 |
| ERR1794913 | Success | 30600 | 0 | 30600 | 1 |
| ERR1794914 | Success | 31282 | 0 | 31282 | 1 |
Antimicrobial Resistance
Per-sample AMR reports
- AMR Report 1 — ERR1794900
- AMR Report 2 — ERR1794901
- AMR Report 3 — ERR1794902
- AMR Report 4 — ERR1794903
- AMR Report 5 — ERR1794904
- AMR Report 6 — ERR1794905
- AMR Report 7 — ERR1794906
- AMR Report 8 — ERR1794907
- AMR Report 9 — ERR1794908
- AMR Report 10 — ERR1794909
- AMR Report 11 — ERR1794910
- AMR Report 12 — ERR1794911
- AMR Report 13 — ERR1794912
- AMR Report 14 — ERR1794913
- AMR Report 15 — ERR1794914
Mobile Genetic Elements
Per-sample MGE reports
- MGE Report 1 — ERR1794900
- MGE Report 2 — ERR1794901
- MGE Report 3 — ERR1794902
- MGE Report 4 — ERR1794903
- MGE Report 5 — ERR1794904
- MGE Report 6 — ERR1794905
- MGE Report 7 — ERR1794906
- MGE Report 8 — ERR1794907
- MGE Report 9 — ERR1794908
- MGE Report 10 — ERR1794909
- MGE Report 11 — ERR1794910
- MGE Report 12 — ERR1794911
- MGE Report 13 — ERR1794912
- MGE Report 14 — ERR1794913
- MGE Report 15 — ERR1794914
Virulence
Per-sample Virulence reports
- Virulence Report 1 — ERR1794909
- Virulence Report 2 — ERR1794910
- Virulence Report 3 — ERR1794911
- Virulence Report 4 — ERR1794912
- Virulence Report 5 — ERR1794913
- Virulence Report 6 — ERR1794914
- Virulence Report 7 — ERR1794900
- Virulence Report 8 — ERR1794901
- Virulence Report 9 — ERR1794902
- Virulence Report 10 — ERR1794903
- Virulence Report 11 — ERR1794904
- Virulence Report 12 — ERR1794905
- Virulence Report 13 — ERR1794906
- Virulence Report 14 — ERR1794907
- Virulence Report 15 — ERR1794908
BLAST
Per-sample BLAST reports
- BLAST Report 1 — ERR1794900
- BLAST Report 2 — ERR1794901
- BLAST Report 3 — ERR1794902
- BLAST Report 4 — ERR1794903
- BLAST Report 5 — ERR1794904
- BLAST Report 6 — ERR1794905
- BLAST Report 7 — ERR1794906
- BLAST Report 8 — ERR1794907
- BLAST Report 9 — ERR1794908
- BLAST Report 10 — ERR1794909
- BLAST Report 11 — ERR1794910
- BLAST Report 12 — ERR1794911
- BLAST Report 13 — ERR1794912
- BLAST Report 14 — ERR1794913
- BLAST Report 15 — ERR1794914
Phylogeny
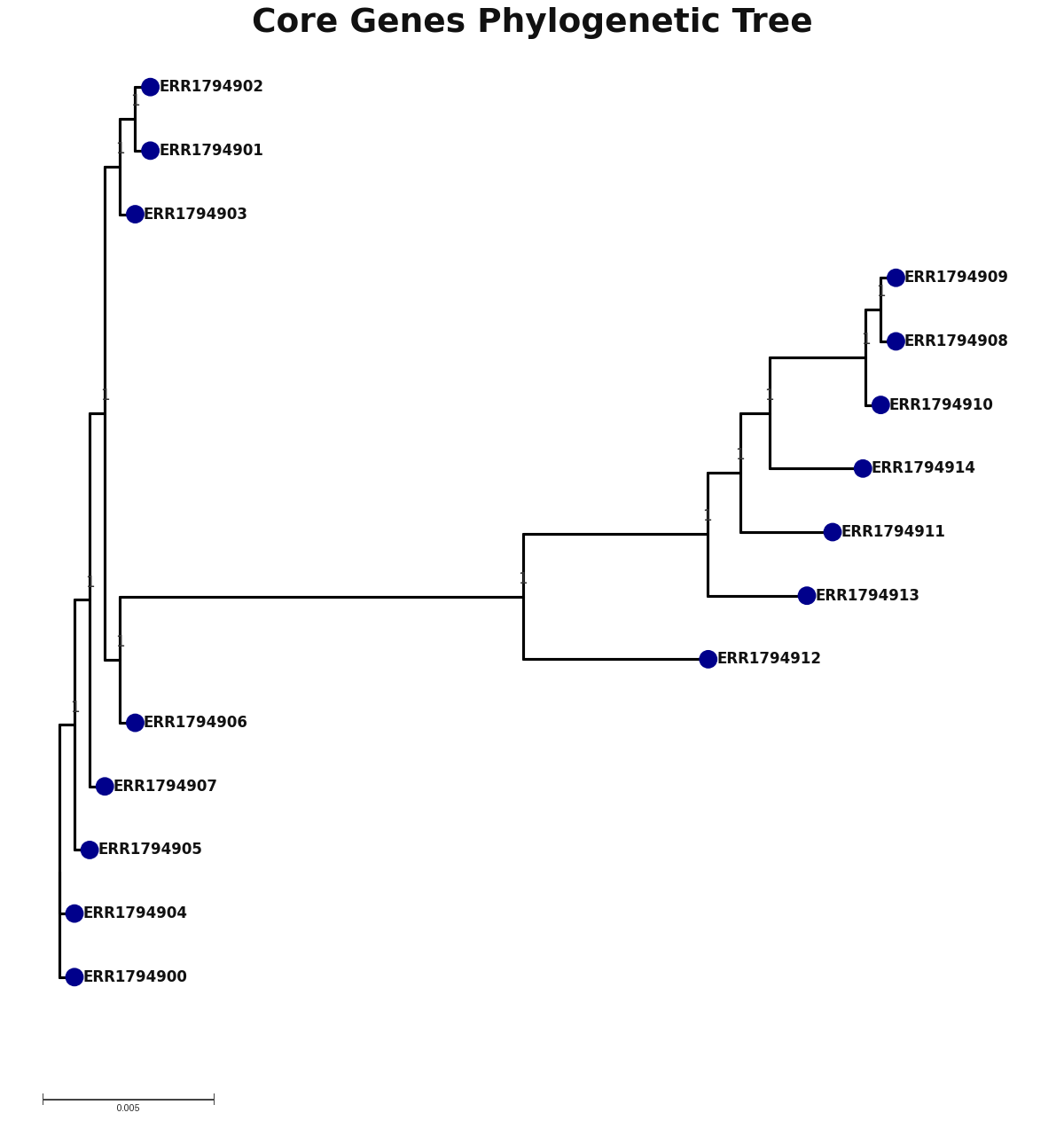
core_core_genes.png
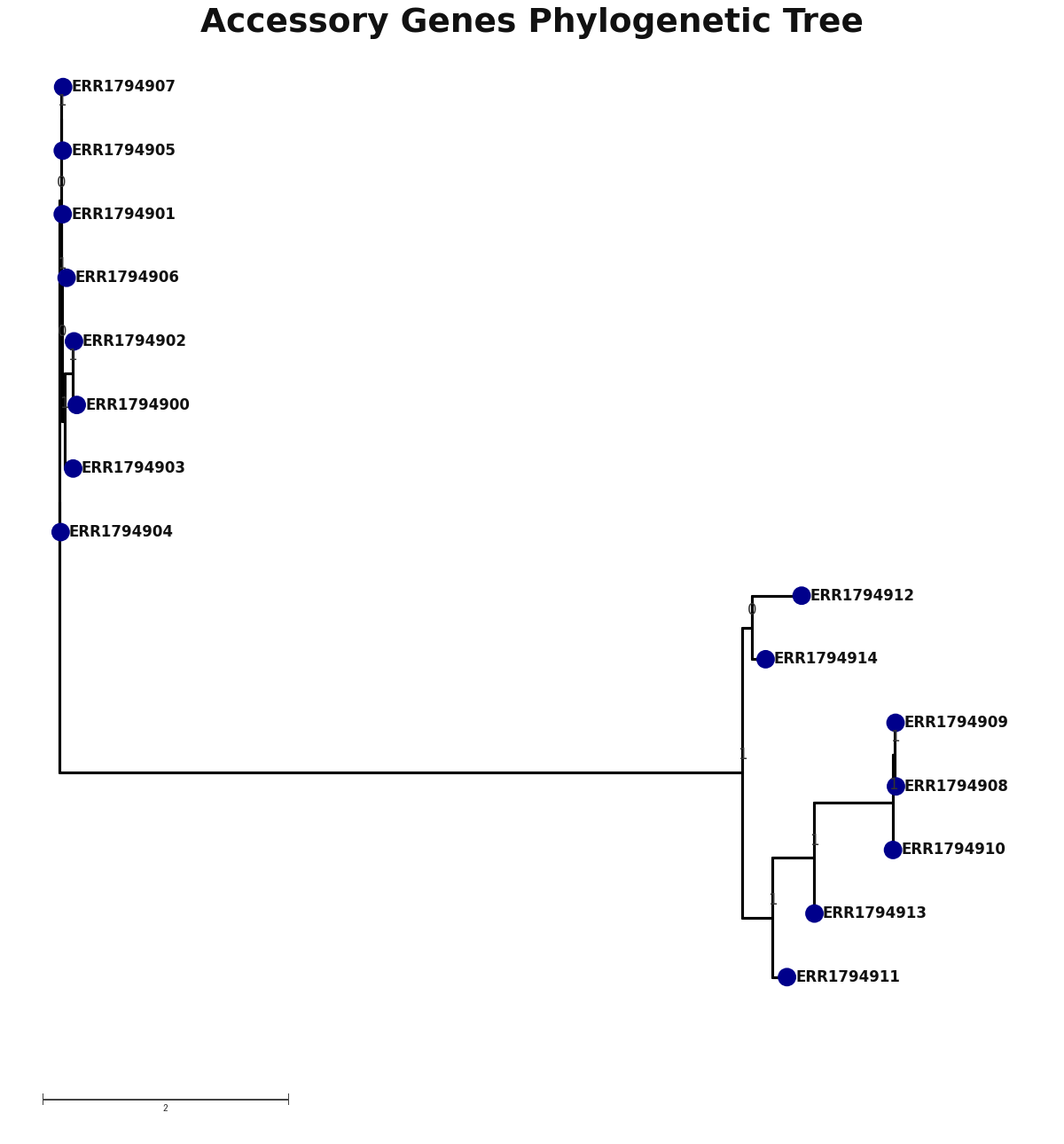
accessory_accessory_genes.png