Analysis Summary
This report aggregates results from rMAP modules (QC, assembly/annotation, pangenome/phylogeny, AMR/MGE/virulence/BLAST, and phylogeny).
Quality Control
Per-sample QC reports
- QC Report 1 — SRR34860533
- QC Report 2 — SRR34940901
- QC Report 3 — SRR34940901
- QC Report 4 — SRR34940902
- QC Report 5 — SRR34940902
- QC Report 6 — SRR34994752
- QC Report 7 — SRR34994752
- QC Report 8 — SRR5514223
- QC Report 9 — SRR5514223
- QC Report 10 — SRR5514224
- QC Report 11 — ERR2397427
- QC Report 12 — SRR5514224
- QC Report 13 — SRR5514226
- QC Report 14 — SRR5514226
- QC Report 15 — SRR5687278
- QC Report 16 — SRR5687278
- QC Report 17 — SRR6208323
- QC Report 18 — SRR6208323
- QC Report 19 — SRR8291573
- QC Report 20 — SRR8291573
- QC Report 21 — SRR8753737
- QC Report 22 — ERR2397427
- QC Report 23 — SRR8753737
- QC Report 24 — SRR8753739
- QC Report 25 — SRR8753739
- QC Report 26 — SRR9029107
- QC Report 27 — SRR9029107
- QC Report 28 — SRR9044171
- QC Report 29 — SRR9044171
- QC Report 30 — SRR9703249
- QC Report 31 — SRR9703249
- QC Report 32 — SRR9964283
- QC Report 33 — SRR10160928
- QC Report 34 — SRR9964283
- QC Report 35 — SRR10160928
- QC Report 36 — SRR10294934
- QC Report 37 — SRR10294934
- QC Report 38 — SRR34860530
- QC Report 39 — SRR34860530
- QC Report 40 — SRR34860533
Read trimming
| Sample | Input Pairs | Both Surviving | Forward Only | Reverse Only | Dropped (%) | Survival Rate (%) |
|---|---|---|---|---|---|---|
| SRR8753739 | 3503310 | 3162430 | 165840 | 81086 | 2.68% | 97.32% |
| ERR2397427 | 1071335 | 888675 | 149170 | 8347 | 2.35% | 97.65% |
| SRR5514226 | 2038819 | 1651949 | 215460 | 65245 | 5.21% | 94.79% |
| SRR5514223 | 2112773 | 1755262 | 182771 | 76397 | 4.65% | 95.35% |
| SRR34940902 | 954929 | 888616 | 33954 | 18908 | 1.41% | 98.59% |
| SRR10294934 | 1860014 | 1633464 | 165744 | 20980 | 2.14% | 97.86% |
| SRR34860530 | 988416 | 911127 | 24314 | 11778 | 4.17% | 95.83% |
| SRR5514224 | 2184225 | 1844365 | 178548 | 68126 | 4.27% | 95.73% |
| SRR8291573 | 3377797 | 3026467 | 179583 | 76551 | 2.82% | 97.18% |
| SRR5687278 | 2701387 | 2337916 | 188255 | 76890 | 3.64% | 96.36% |
| SRR34860533 | 1013425 | 932873 | 25445 | 11584 | 4.29% | 95.71% |
| SRR9029107 | 4859583 | 4413497 | 230856 | 101210 | 2.35% | 97.65% |
| SRR10160928 | 574518 | 515652 | 34899 | 8551 | 2.68% | 97.32% |
| SRR34940901 | 842378 | 731548 | 61246 | 20383 | 3.47% | 96.53% |
| SRR6208323 | 442192 | 423416 | 11747 | 657 | 1.44% | 98.56% |
| SRR9044171 | 746958 | 643548 | 64790 | 7496 | 4.17% | 95.83% |
| SRR8753737 | 3036843 | 2698744 | 168437 | 87779 | 2.70% | 97.30% |
| SRR9703249 | 551969 | 509394 | 34192 | 2341 | 1.09% | 98.91% |
| SRR9964283 | 608692 | 532989 | 55551 | 4855 | 2.51% | 97.49% |
| SRR34994752 | 944334 | 804132 | 106309 | 10066 | 2.52% | 97.48% |
Assembly
Assembly statistics
| Sample | Contigs | Total Length | Min Length | Max Length | Avg Length | N50 |
|---|---|---|---|---|---|---|
| ERR2397427 | 1566 | 5879148 | 78 | 356894 | 3754 | 126797 |
| SRR10160928 | 772 | 5897517 | 78 | 259762 | 7639 | 86409 |
| SRR10294934 | 1422 | 5739820 | 78 | 306299 | 4036 | 134242 |
| SRR34860530 | 396 | 5493958 | 78 | 272482 | 13873 | 140755 |
| SRR34860533 | 526 | 5609587 | 78 | 350426 | 10664 | 106020 |
| SRR34940901 | 554 | 5613931 | 78 | 384183 | 10133 | 109595 |
| SRR34940902 | 489 | 5624375 | 78 | 344268 | 11501 | 100595 |
| SRR34994752 | 907 | 5433932 | 78 | 373451 | 5991 | 114897 |
| SRR5514223 | 712 | 5608645 | 78 | 276256 | 7877 | 94157 |
| SRR5514224 | 883 | 5674283 | 78 | 323272 | 6426 | 112735 |
| SRR5514226 | 664 | 5440085 | 78 | 272540 | 8192 | 119267 |
| SRR5687278 | 1373 | 5652038 | 78 | 225828 | 4116 | 123455 |
| SRR6208323 | 1103 | 6158839 | 78 | 220899 | 5583 | 54129 |
| SRR8291573 | 511 | 5605274 | 78 | 269057 | 10969 | 133039 |
| SRR8753737 | 12021 | 7930530 | 78 | 348129 | 659 | 70623 |
| SRR8753739 | 11973 | 7572699 | 78 | 219741 | 632 | 65439 |
| SRR9029107 | 902 | 5585611 | 78 | 287592 | 6192 | 96574 |
| SRR9044171 | 971 | 5980249 | 78 | 244308 | 6158 | 95517 |
| SRR9703249 | 1053 | 5342066 | 78 | 356066 | 5073 | 114515 |
| SRR9964283 | 906 | 5333384 | 78 | 286455 | 5886 | 106527 |
Annotation
Annotation summary
| Sample ID | Gene Count |
|---|---|
| ERR2397427 | 5327 |
| SRR10160928 | 5529 |
| SRR10294934 | 5194 |
| SRR34860530 | 5016 |
| SRR34860533 | 5107 |
| SRR34940901 | 5176 |
| SRR34940902 | 5194 |
| SRR34994752 | 4921 |
| SRR5514223 | 5167 |
| SRR5514224 | 5238 |
| SRR5514226 | 5005 |
| SRR5687278 | 5110 |
| SRR6208323 | 5775 |
| SRR8291573 | 5175 |
| SRR8753737 | 5862 |
| SRR8753739 | 5531 |
| SRR9029107 | 5113 |
| SRR9044171 | 5476 |
| SRR9703249 | 5024 |
| SRR9964283 | 4901 |
Pangenome
Pangenome report
Pangenome Analysis
Roary completed successfully. (This page will show Roary results if available.)

Gene Presence/Absence Heatmap
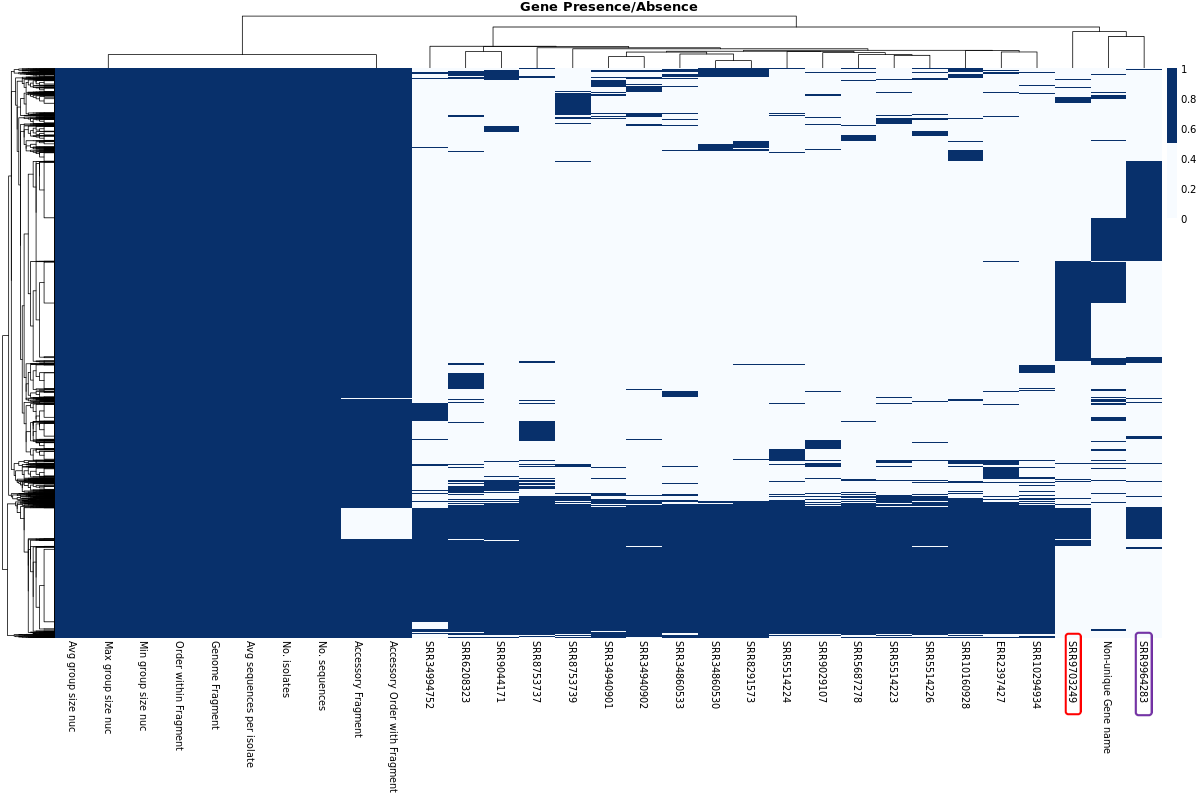
MLST
Combined MLST
| Sample ID | Species | ST | gapA | infB | mdh | pgi | phoE | rpoB | tonB |
|---|---|---|---|---|---|---|---|---|---|
| ERR2397427 | kpneumoniae | 35 | 2 | 1 | 2 | 1 | 10 | 1 | 19 |
| SRR10160928 | kpneumoniae | 893 | 25 | 1 | 101 | 1 | 10 | 1 | 100 |
| SRR10294934 | kpneumoniae | 231 | 2 | 6 | 1 | 3 | 26 | 1 | 77 |
| SRR34860530 | kpneumoniae | 23 | 2 | 1 | 1 | 1 | 9 | 4 | 12 |
| SRR34860533 | kpneumoniae | 65 | 2 | 1 | 2 | 1 | 10 | 4 | 13 |
| SRR34940901 | kpneumoniae | 2039 | 2 | 3 | 2 | 4 | 9 | 4 | 13 |
| SRR34940902 | kpneumoniae | 66 | 2 | 3 | 2 | 1 | 10 | 1 | 13 |
| SRR34994752 | kpneumoniae | - | 17 | 19 | 156 | ~39 | ~207 | 21 | 589 |
| SRR5514223 | kpneumoniae | 48 | 2 | 5 | 2 | 2 | 7 | 1 | 10 |
| SRR5514224 | kpneumoniae | 1263 | 4 | 1 | 1 | 1 | 12 | 1 | 35 |
| SRR5514226 | kpneumoniae | 530 | 2 | 39 | 2 | 1 | 1 | 4 | 24 |
| SRR5687278 | kpneumoniae | 4 | 3 | 1 | 1 | 1 | 3 | 3 | 1 |
| SRR6208323 | kpneumoniae | 15 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| SRR8291573 | kpneumoniae | 23 | 2 | 1 | 1 | 1 | 9 | 4 | 12 |
| SRR8753737 | kpneumoniae | 37 | 2 | 9 | 2 | 1 | 13 | 1 | 16 |
| SRR8753739 | kpneumoniae | 2121 | 3 | 1 | 1 | 4 | 7 | 4 | 13 |
| SRR9029107 | kpneumoniae | 1798 | 3 | 5 | 1 | 134 | 10 | 4 | 4 |
| SRR9044171 | kpneumoniae | 14 | 1 | 6 | 1 | 1 | 1 | 1 | 1 |
| SRR9703249 | ecloacae | 182 | 49 | 20 | 19 | 44 | 90 | 24 | 32 |
| SRR9964283 | ecoli | 131 | 53 | 40 | 47 | 13 | 36 | 28 | 29 |
Variants
Variant summary
| Sample | Status | SNPs | INDELs | Total Variants | Attempts |
|---|---|---|---|---|---|
| ERR2397427 | Success | 25542 | 0 | 25542 | 1 |
| SRR10160928 | Success | 23346 | 0 | 23346 | 1 |
| SRR10294934 | Success | 25417 | 0 | 25417 | 1 |
| SRR34860530 | Success | 25075 | 0 | 25075 | 1 |
| SRR34860533 | Success | 24850 | 0 | 24850 | 1 |
| SRR34940901 | Success | 24730 | 0 | 24730 | 1 |
| SRR34940902 | Success | 24819 | 0 | 24819 | 1 |
| SRR34994752 | Success | 131368 | 0 | 131368 | 1 |
| SRR5514223 | Success | 23987 | 0 | 23987 | 1 |
| SRR5514224 | Success | 23529 | 0 | 23529 | 1 |
| SRR5514226 | Success | 23920 | 0 | 23920 | 1 |
| SRR5687278 | Success | 25992 | 0 | 25992 | 1 |
| SRR6208323 | Success | 23855 | 0 | 23855 | 1 |
| SRR8291573 | Success | 25488 | 0 | 25488 | 1 |
| SRR8753737 | Success | 24437 | 0 | 24437 | 1 |
| SRR8753739 | Success | 24438 | 0 | 24438 | 1 |
| SRR9029107 | Success | 24543 | 0 | 24543 | 1 |
| SRR9044171 | Success | 25279 | 0 | 25279 | 1 |
| SRR9703249 | Success | 30350 | 0 | 30350 | 1 |
| SRR9964283 | Success | 16184 | 0 | 16184 | 1 |
Antimicrobial Resistance
Per-sample AMR reports
- AMR Report 1 — ERR2397427
- AMR Report 2 — SRR10160928
- AMR Report 3 — SRR10294934
- AMR Report 4 — SRR34860530
- AMR Report 5 — SRR34860533
- AMR Report 6 — SRR34940901
- AMR Report 7 — SRR34940902
- AMR Report 8 — SRR34994752
- AMR Report 9 — SRR5514223
- AMR Report 10 — SRR5514224
- AMR Report 11 — SRR5514226
- AMR Report 12 — SRR5687278
- AMR Report 13 — SRR6208323
- AMR Report 14 — SRR8291573
- AMR Report 15 — SRR8753737
- AMR Report 16 — SRR8753739
- AMR Report 17 — SRR9029107
- AMR Report 18 — SRR9044171
- AMR Report 19 — SRR9703249
- AMR Report 20 — SRR9964283
Mobile Genetic Elements
Per-sample MGE reports
- MGE Report 1 — ERR2397427
- MGE Report 2 — SRR10160928
- MGE Report 3 — SRR10294934
- MGE Report 4 — SRR34860530
- MGE Report 5 — SRR34860533
- MGE Report 6 — SRR34940901
- MGE Report 7 — SRR34940902
- MGE Report 8 — SRR34994752
- MGE Report 9 — SRR5514223
- MGE Report 10 — SRR5514224
- MGE Report 11 — SRR5514226
- MGE Report 12 — SRR5687278
- MGE Report 13 — SRR6208323
- MGE Report 14 — SRR8291573
- MGE Report 15 — SRR8753737
- MGE Report 16 — SRR8753739
- MGE Report 17 — SRR9029107
- MGE Report 18 — SRR9044171
- MGE Report 19 — SRR9703249
- MGE Report 20 — SRR9964283
Virulence
Per-sample Virulence reports
- Virulence Report 1 — SRR5514224
- Virulence Report 2 — SRR5514226
- Virulence Report 3 — SRR5687278
- Virulence Report 4 — SRR6208323
- Virulence Report 5 — SRR8291573
- Virulence Report 6 — SRR8753737
- Virulence Report 7 — SRR8753739
- Virulence Report 8 — SRR9029107
- Virulence Report 9 — SRR9044171
- Virulence Report 10 — SRR9703249
- Virulence Report 11 — ERR2397427
- Virulence Report 12 — SRR9964283
- Virulence Report 13 — SRR10160928
- Virulence Report 14 — SRR10294934
- Virulence Report 15 — SRR34860530
- Virulence Report 16 — SRR34860533
- Virulence Report 17 — SRR34940901
- Virulence Report 18 — SRR34940902
- Virulence Report 19 — SRR34994752
- Virulence Report 20 — SRR5514223
BLAST
Per-sample BLAST reports
- BLAST Report 1 — ERR2397427
- BLAST Report 2 — SRR10160928
- BLAST Report 3 — SRR10294934
- BLAST Report 4 — SRR34860530
- BLAST Report 5 — SRR34860533
- BLAST Report 6 — SRR34940901
- BLAST Report 7 — SRR34940902
- BLAST Report 8 — SRR34994752
- BLAST Report 9 — SRR5514223
- BLAST Report 10 — SRR5514224
- BLAST Report 11 — SRR5514226
- BLAST Report 12 — SRR5687278
- BLAST Report 13 — SRR6208323
- BLAST Report 14 — SRR8291573
- BLAST Report 15 — SRR8753737
- BLAST Report 16 — SRR8753739
- BLAST Report 17 — SRR9029107
- BLAST Report 18 — SRR9044171
- BLAST Report 19 — SRR9703249
- BLAST Report 20 — SRR9964283
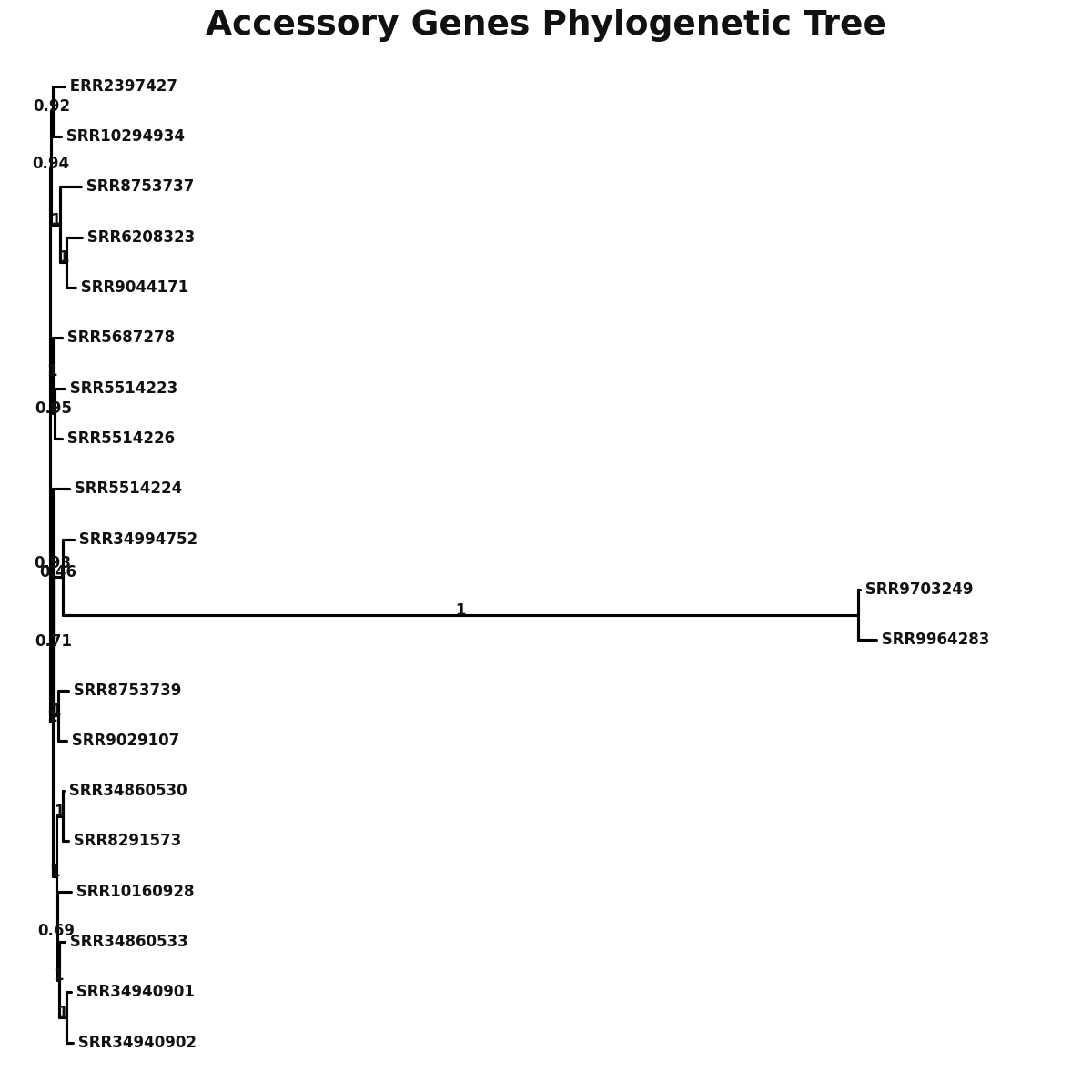
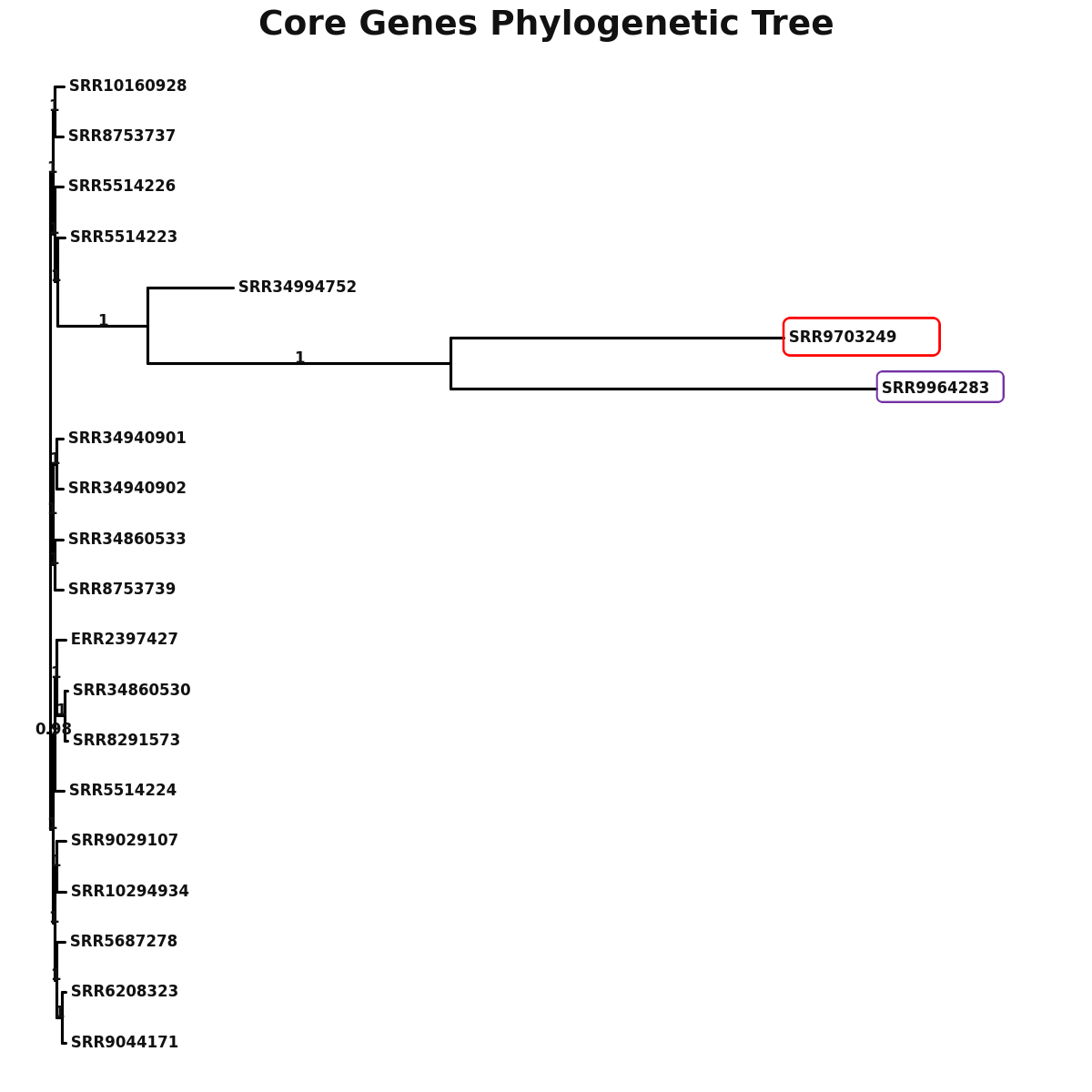
Phylogeny
core_core_genes.png
accessory_accessory_genes.png